10 Top Tips to Help Reduce the Cost of Your NGS Workflow
Next-generation sequencing (NGS) has lowered the cost and time required for sequencing, making genomic-level insights more accessible to academia and industry. Here are our 10 top tips to help you further reduce the cost of your NGS workflows.
DNA sequencing has come a long way since the completion of the human genome project, and next-generation sequencing (NGS) is now an essential tool in the modern laboratory. However, NGS still has considerable costs attached to it. Sample and library preparation, sequencing itself, data analysis, and the terabytes or more of data storage are all sources of cost and inefficiencies. So, how can you keep your costs down and improve both scalability and reliability?
In this list, you’ll find our ten top tips for reducing costs in your NGS workflows, whether you only sequence occasionally or run a series of high-throughput platforms.
1. Start Using NGS
This might seem like an odd tip to start with, but Sanger sequencing is still in heavy use across academia and industry, and many might not have considered trying NGS yet.
Sanger sequencing has its purposes. If you need to check the integration of a short vector or gene editing insert, sequence a single gene, or perform some basic microsatellite analysis, Sanger sequencing is the way to go.
For anything more substantial, such as the sequencing of a whole chromosome or genome or identifying novel variants, NGS provides a far more comprehensive data output. Amplicon sequencing on small NGS platforms can be an ideal stepping stone where your throughput allows.
NGS opens new possibilities for experimental design and, as outlined in a later tip, does not require you to have a lot of specialist knowledge to get started, and it can have a considerably lower costper- kilobase sequenced than Sanger sequencing.
2. Try Third-Party Consumables for NGS
Sequencing services and platform vendors are likely to recommend specific (often their own) kits for sample and library preparation. They might claim these kits are optimized for or validated on their platforms, however, the ubiquitous nature and understanding of NGS technologies now means third-party consumables can be both more cost-effective and perform just as well or better than vendor kits.
Third-party kits provide you with flexibility for experimental design, and having the ability to shop around for the best performance, workflows, and deals means you can improve efficiency while keeping costs down. For example, rather than separate reagents for isolation, size selection, and PCR clean-up—key steps in sample and library preparation—you could use magnetic bead-based kits to have a single solution and automatable workflow.
3. Magnetize your Sample (and Library) Preparation
 Speaking of magnetic beads, it is worth highlighting just how versatile these super (paramagnetic) little particles are in the NGS workflow. Magnetic beads are well suited to DNA extraction, size selection, and clean-up as alternatives to column based purification, providing flexibility in scale and handling.
Speaking of magnetic beads, it is worth highlighting just how versatile these super (paramagnetic) little particles are in the NGS workflow. Magnetic beads are well suited to DNA extraction, size selection, and clean-up as alternatives to column based purification, providing flexibility in scale and handling.
This approach can greatly simplify extraction and size selection steps while introducing a level of consistency that is challenging to achieve otherwise between different staff and labs. Magnetic beads can also simplify library preparation for multiplexed sequencing and are readily automatable. Find out more about these in the tips below.
4. Amplify with Phi
NGS workflows can be sample-hungry, fraught with variation, and create challenges when your sample is precious and in limited supply. In many cases, you should consider a sampleamplification step as essential for doing the required analysis and also providing security in case repeats are required.
The traditional approach, PCR, often requires optimization. Primers bind some areas of the genome more efficiently than others. Thermal cycling can introduce bias and vary in efficiency depending on the quality of the reagents and the thermal cycler you use.
Phi29 polymerase provides reliable and isothermal amplification of circular and linear DNA using random primers. A short reaction at 30°C can amplify the picograms of DNA from a single cell to micrograms for sequencing, generating fragments often over 70 kb in size.
This high-fidelity and highly processive polymerase removes a considerable amount of doubt in the amplification process, and minimizes the reliance on PCR to make the amplification step more reliable and efficient.
5. Automate your NGS Workflow
 Automation is not just for large high throughput sequencing facilities. The entry costs for automation are lower than they have ever been, and basic liquid handling systems need not take more space than a laminar flow hood.
Automation is not just for large high throughput sequencing facilities. The entry costs for automation are lower than they have ever been, and basic liquid handling systems need not take more space than a laminar flow hood.
Automating these basic tasks both reduces your hands-on time, freeing up members of your team for other projects, improves the overall speed of the workflow, and reduces the chances of errors.
6. Track your Samples Automatically (or at least easily)
 Like automation, using barcodes to track and maintain a database of samples and their properties is no longer just for large facilities. The result of mixing up two samples or incorrectly labeling data is just as concerning for the academic researcher as it is for sequencing service providers.
Like automation, using barcodes to track and maintain a database of samples and their properties is no longer just for large facilities. The result of mixing up two samples or incorrectly labeling data is just as concerning for the academic researcher as it is for sequencing service providers.
Hand-labeling samples and noting their specifics in a notebook might work well enough for a few tubes. But as batch and database sizes increase, there is a likelihood of introducing errors. These errors then lead to staff collecting, preparing, and sequencing samples again, an unnecessary waste of time and resources.
Barcodes and readers are readily available and reduce the risks of cross contamination and sample mix-ups. Many entry-level plate handling and storage systems also incorporate barcode readers, so a modestly-sized facility can also benefit from automatic sample tracking.
7. Scale Down Volumes to Scale Up Numbers and Cut Costs
It is common to work in the 96-well format, especially when handling samples manually. An eight-way pipette and a steady hand makes preparing the occasional plate of samples for sequencing quick and easy.
However, if you tend to have larger batches of samples, and might already be considering automation, it could also be worth switching to a 384-well format. Not only will the smaller format reduce the number of plates and reagent necessary, but combine it with automation and you could process samples with less variation and waste.
8. Multiplexing Need not be Perplexing
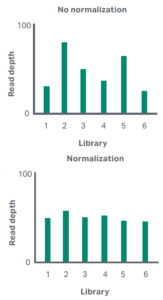 In Sanger sequencing, one run equals one short sequence. In NGS, indexing adapters or molecular barcodes means one run can equal hundreds or even thousands of libraries of sequences.
In Sanger sequencing, one run equals one short sequence. In NGS, indexing adapters or molecular barcodes means one run can equal hundreds or even thousands of libraries of sequences.
Multiplexing like this is what has driven down the cost of sequencing. It enables you to run multiple libraries on a single flow cell, making maximum use of the capacity of your NGS platform and so make it more cost-effective.
Multiplexing is not without its challenges though, one of which is the normalization of NGS libraries.
DNA normalization is the process of equalizing the concentration of DNA libraries for multiplexing so that no library is over- or under-represented in the sequencing output.
Usually, the process involves careful measurement and dilution of the libraries, but once again, magnetic beads can provide a remarkably simple option that actually does away with the quantitation step altogether.
9. Know when to Scale Up and Scale Down
Increasing parallel sequencing capacity has been essential in reducing the costs of NGS. In addition to multiplexing, some sequencing platforms provide you with the option to run multiple flow cells, effectively multiplying your sequencing capacity without also multiplying your costs.
On the other hand, when your needs are more modest, these large high-throughput sequencing platforms can seem like a substantial investment with a capacity far beyond your current requirements.
That is where small benchtop sequencing platforms can make more sense. These sequencers can integrate easily into your lab and are well suited for whole-genome sequencing of microbes and viruses, as well as targeted gene sequencing.
10. Outsource Sequencing and Storage
Ultimately, though, it is worth asking your team whether they need in-house sequencing right now. Any sequencing platform is a long-term investment and will involve upkeep and optimization.
As an alternative, you could take advantage of the many sequencing service providers out there. They will have established and optimized workflows that turn your prepared samples into reliable data.
You could also reduce the burden of bioinformatics by using ready-made cloud apps to look after your projects and process your data.
And what about all that data? You could potentially store terabytes of data locally, but an off-site backup is essential for security and redundancy. This might be in the form of cloud-based storage for recent or regularly-accessed data, or tape-based cold storage for archiving data.
Those same sequencing providers might also offer dedicated NGS data storage options, which could be a cost-effective option as they will already have their own data safeguarding measures in place, saving you the effort.
Want More?
Sign up for the GSS Resources Newsletter, an eNewsletter that delivers new technology and news, straight to your inbox.
GSS-Sales@govsci.com | 800.248-8030 | govsci.com

ISO 9001:2015 Certified
Copyright 2020, Government Scientific Source. All Rights Reserved.







