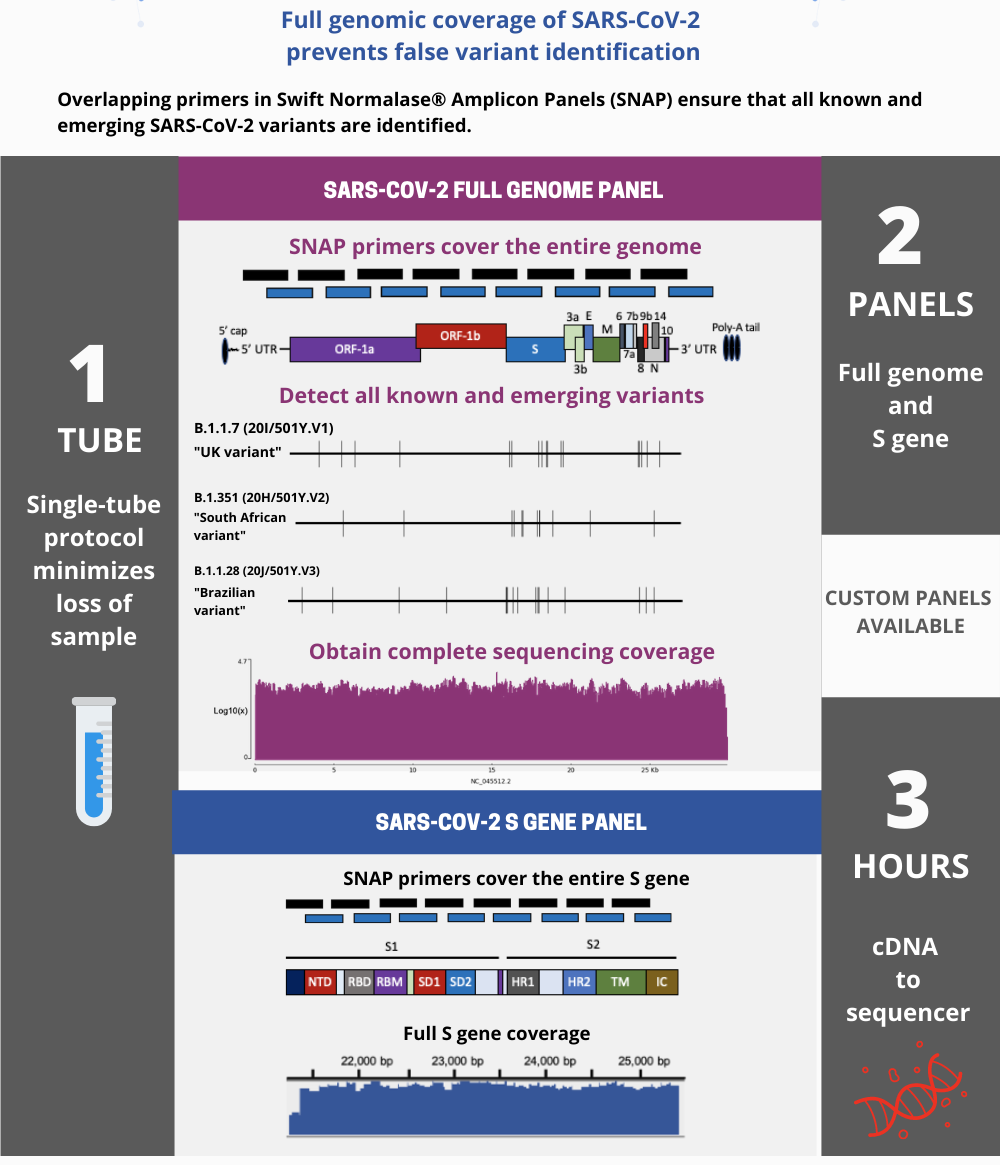
Detect COVID-19 Variants with Swift Biosciences’ SARS CoV-2 Panel
Full Genomic Coverage Obtains Genomes from All Variants
Obtain genomes from viral titers as low as 10-100 viral copies
10 to 1 million viral genome copies are sufficient to generate NGS libraries using the SNAP panels. Following detection of SARS-CoV-2 by qPCR, the remaining cDNA is used as input for the SNAP Panels.
Two SNAP Panels are available to sequence SARS-CoV-2
Obtain full coverage of SARS-CoV-2 genome
The SNAP SARS-CoV-2 Kit uses overlapping primers to generate 345 amplicons, sized 116-255 bp (average 150 bp), along the length of the 29.9 kb viral genome and obtain 99.7% coverage of the genome. Overlapping primers ensure that novel variants are detected, even when the mutation is a dropout of genomic sequence. Comprehensive mutation detection is crucial for tracking nucleotide variants and improve understanding of virus evolution, transmission, and pathogenesis.
Obtain full coverage of S Gene within SARS-CoV-2 genome
The Swift Normalase Amplicon SARS-CoV-2 S Gene NGS Panel (SNAP) provides 100% coverage of the gene which encodes for the Spike Protein in the virus which causes COVID-19. Identify presence of emerging viral strains which contain mutations in the S Gene and model potential changes in transmission patterns based upon those mutations.
If variants are there, they will been seen.
See full details at: https://swiftbiosci.com/swift-amplicon-sars-cov-2-research-panels/
This product is for Research Use Only. Not for use in diagnostic procedures.
Want More?
Sign up for the GSS Resources Newsletter, an eNewsletter that delivers new technology and news, straight to your inbox.
GSS-Sales@govsci.com | 800.248-8030 | govsci.com

ISO 9001:2015 Certified
Copyright 2020, Government Scientific Source. All Rights Reserved.







